Research
My journey in the scientific world has been largely centered around a fascinating technology known as Nanopore DNA sequencing. I often describe this technology as a 'DNA-scanner.' Given the minuscule size of DNA, about 100,000 times smaller in diameter than a human hair, the scanner must also be incredibly small, hence the 'nano'-pore!
I am proud to have played a significant role in the development of the first functional technology of this kind, making several fundamental contributions. My work continues in the industry as I coordinate teams and contribute to the development of the Machine Learning that unlocks the full potential of this technology.
For a more detailed look into my journey and the intricacies of genetic information flow with next-gen sequencers, check out my Bio info story. I've also included a list of my academic publications. Feel free to reach out if you have any questions.
Research Publications¶
Nanopore tweezers show fractional-nucleotide translocation in sequence-dependent pausing by RNA polymerase¶
With Single-Molecule Picometer Resolution Nanopore Tweezers we measure how a RNA polymerase moves with exquisite detail
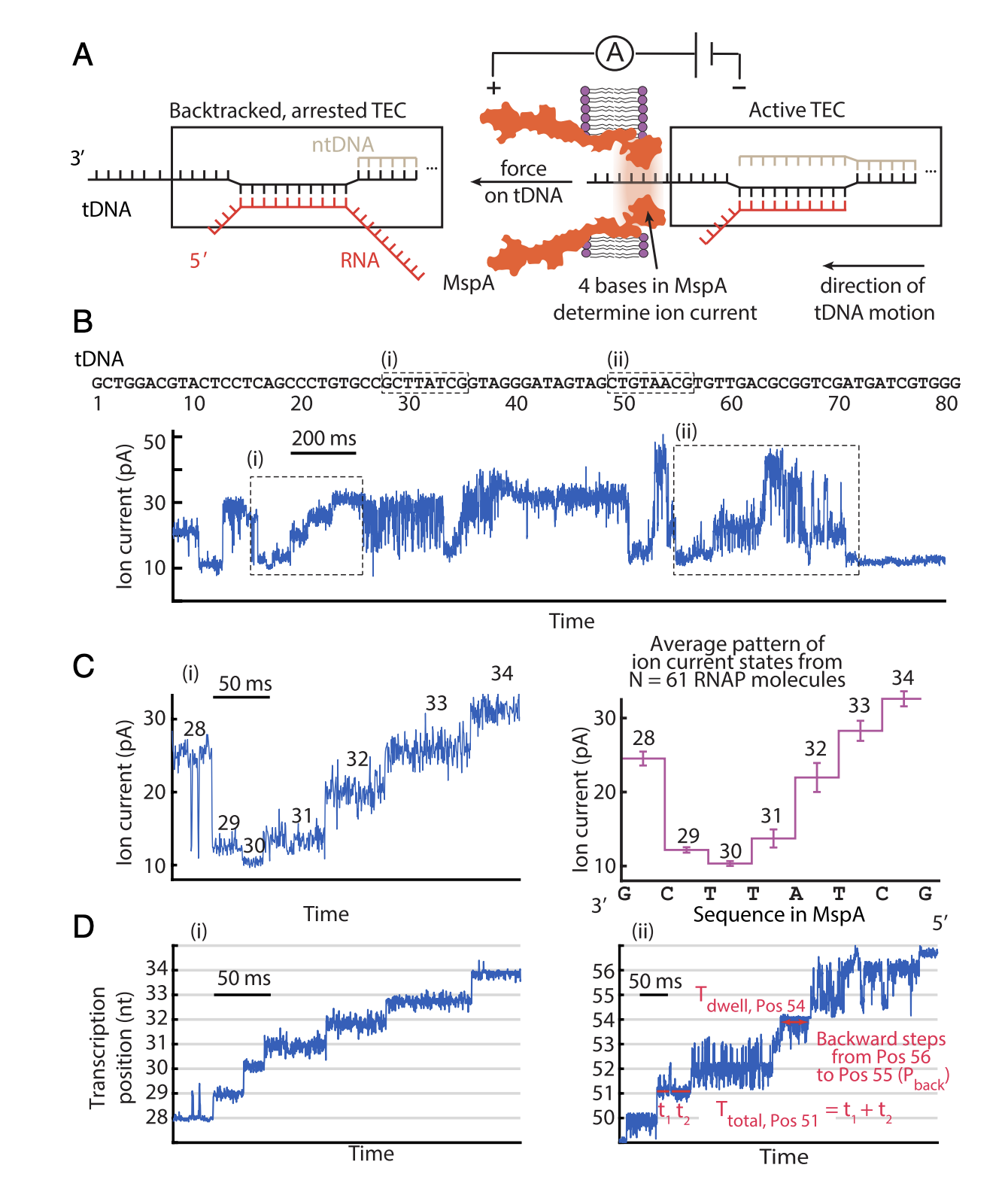
Ian C. Nova, Jonathan M. Craig, Abhishek Mazumder, Andrew H. Laszlo, Ian M. Derrington, Matthew T. Noakes, Henry Brinkerhoff, Shuya Yang, Hanif Vahedian-Movahed, Lingting Li, Yu Zhang, Jasmine L. Bowman, Jesse R. Huang, Jonathan W. Mount, Richard H. Ebright, and Jens H. Gundlach
Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane (2019)¶
Use DNA's inherent flexibility to create a more 'continuous scan' of the DNA as it moves through the nanopore to increase sequencing accuracy.
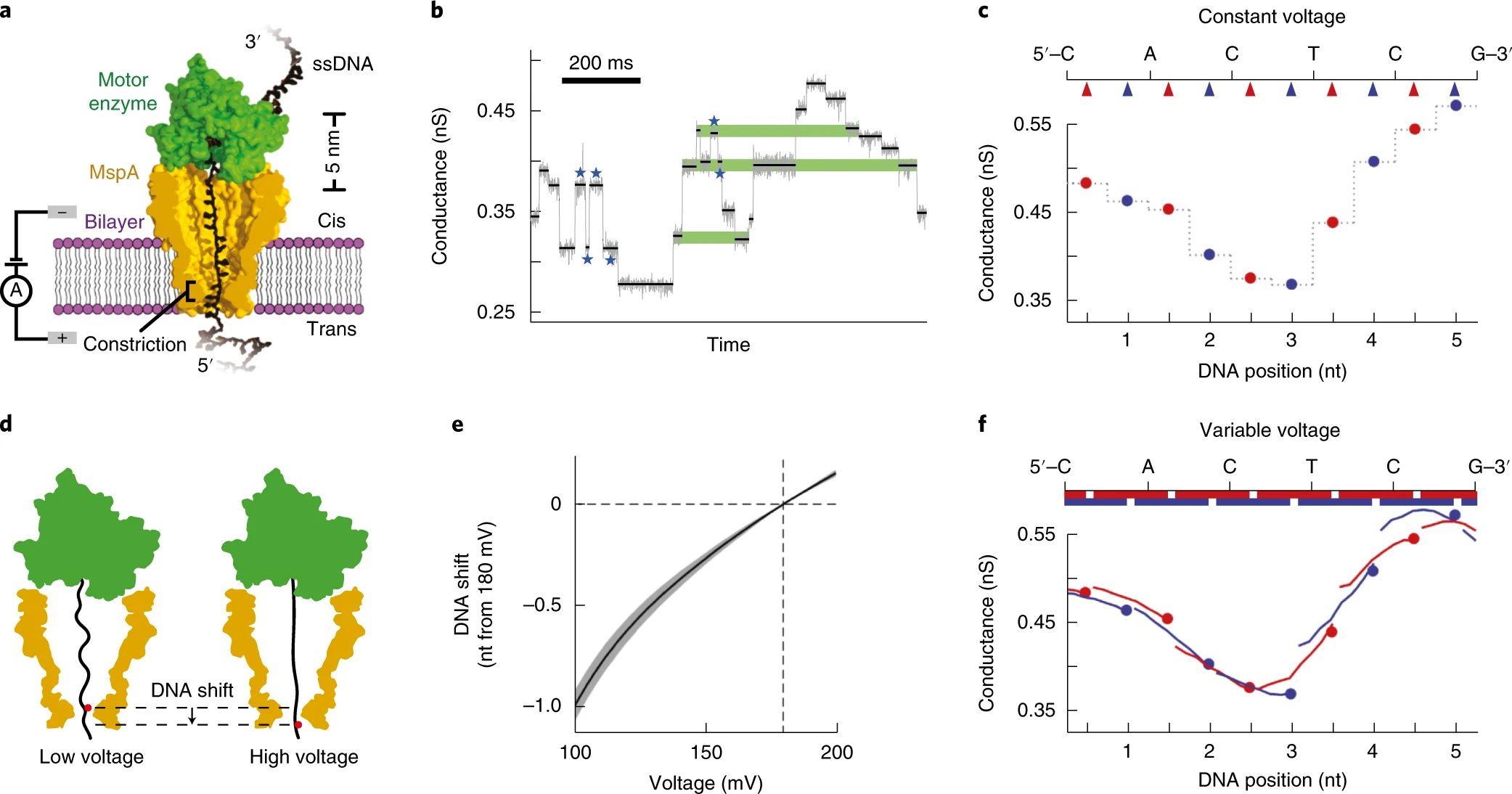
M.T. Noakes, H. Brinkerhoff, A.H. Laszlo, I.M. Derrington, K.W. Langford, J.W. Mount, J.L. Bowman, K.M. Doering, B.I.Tickman, J.H. Gundlach.
Nature Biotechnology. doi: 10.1038/s41587-019-0096-0 doi: 10.1038/s41587-019-0096-0
Single Molecule DNA detection with an engineered MspA protein nanopore (2008)¶
We engineered the nanopore MspA and demonstrated that this enables DNA to move through it.
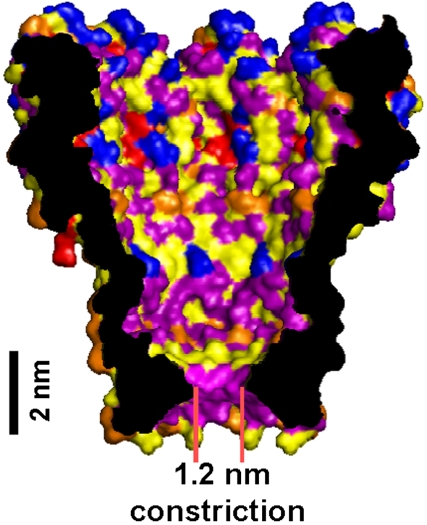
T. Butler, M. Pavlenok, I.M. Derrington, M. Niederweis, and J. Gundlach.
Subangstrom Measurements of Enzyme Function Using a Biological Nanopore, SPRNT (2017)¶
Using the exquisite sub-angstrom precision of the MspA nanopore, we directly measured sub-catalysis reaction steps for the Hel-308 helicase
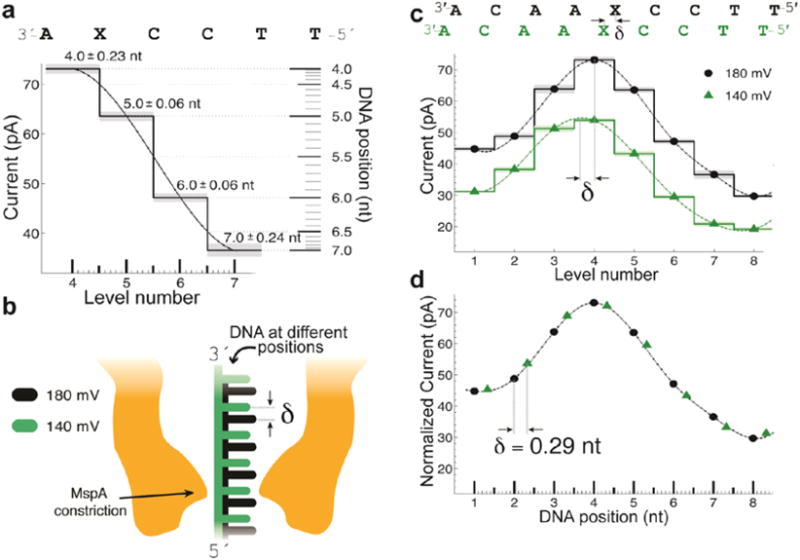
Using the exquisite sub-angstrom precision of the MspA nanopore, we directly measured sub-catalysis reaction steps for the Hel-308 helicase, providing a greater understanding of the mechanism of the superfamily 2 helicase.
Laszlo A.H., I.M. Derrington, J.H. Gundlach.
Reading DNA with the MspA Nanopore and a Motor Enzyme (2012)¶
We demonstrated for the first time enzyme-controlled motion of DNA through a nanopore, directly demonstrating the potential for high-throughput nanopore-sequencing.
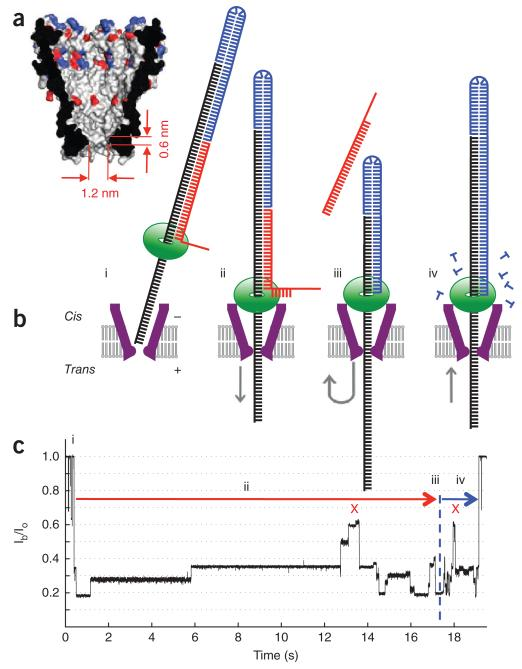
E. Manrao, I.M. Derrington, A. Laszlo, K. Langford, M. Hopper, N. Gilgren, M. Pavlenok, M. Niederweis, J. Gundlach.
Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA (2013)¶
We demonstrated that the important epigenetic marker of DNA methylation is easily detectable with sequencing using the nanopore MspA.
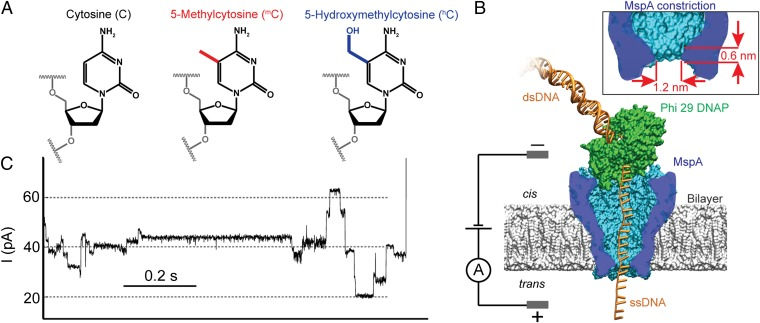
A. Laszlo, I. M. Derrington, H. Brinkerhoff, K. Langford, I. Nova, J. Samson, J. Bartlett, M. Pavlenok, J. Gundlach.
Subangstrom single-molecule measurements of motor proteins using a nanopore (2015)¶
We demonstrated that the nanopore MspA can be used to resolve the position of DNA at sub-angstrom values at bandwidths of greater than a kHz, providing the basis of a novel tool to understand enzymes.
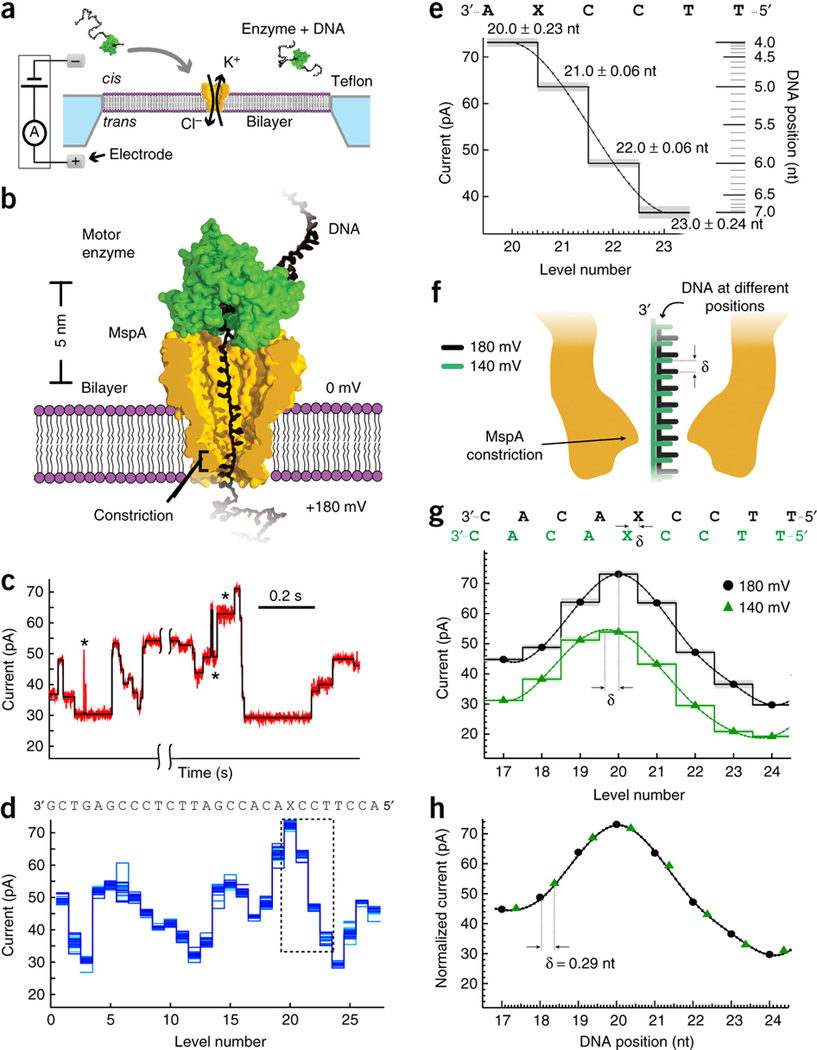
I.M. Derrington, J.M. Craig, E.Stava, A.H. Laszlo, B.C. Ross, H. Brinkerhoff, I.C. Nova, K. Doering, B. Tickman, M. Ronaghi, J.G. Mandell, K.L. Gunderson, J.H. Gundlach.
Nanopore DNA sequencing with MspA (2010)¶
We showed that the Nanopore MspA has the requisite sensitivity to sequence DNA and provided a method to potentially enable sequencing with it.
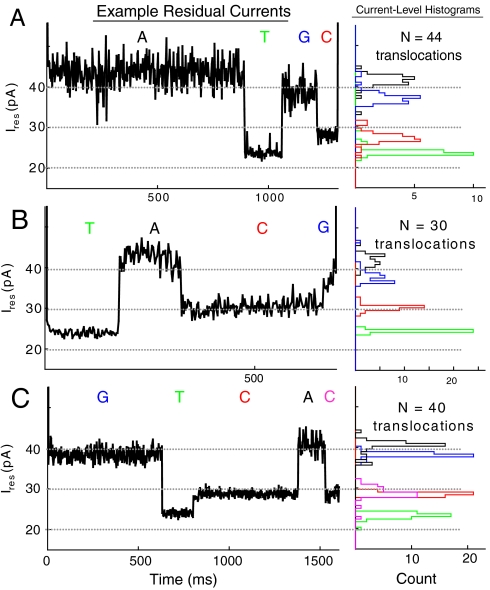
I.M. Derrington, M. Collins, E. Manrao, M. Pavlenok, M. Niederweis, J.H. Gundlach.
Decoding long nanopore sequencing reads of natural DNA (2014)¶
With the understood ability to sequence with the nanopore MspA, we demonstrated, for the first time, multi-kb reads of genomic DNA.
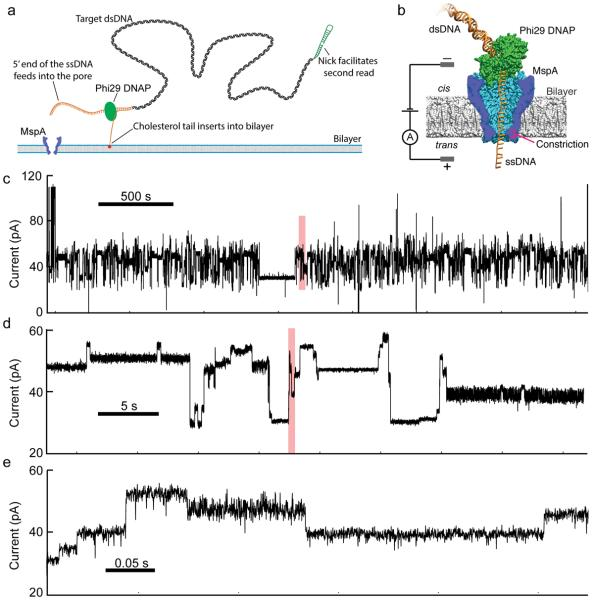
A.H. Laszlo, I. M. Derrington, B.C. Ross, H. Brinkerhoff, A. Adey, I.C. Nova, J.M. Craig, K.W. Langford, J.M. Samson, R. Daza, K. Doering, J. Shendure, J.H. Gundlach.